| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Power Spectrum Analysis Up: Angular Distribution Functions Previous: Angular Distribution Functions
Syntax
- DYNAmics
- ANALysis ADF {
 dynamics-adf-statement
dynamics-adf-statement } END is invoked
from the main level of X-PLOR.
} END is invoked
from the main level of X-PLOR.
 dynamics-adf-statement
dynamics-adf-statement :==
:==-
 trajectory-statement
trajectory-statement
- defines the input data; see Section 12.1.3.
- AGRId=
 real
real
- specifies an angular grid (default: 10.0).
- ASPEcies=
 selection
selection
- selects “A"-species.
- BSPEcies=
 selection
selection
- selects “B"-species.
- DENSity=
 real
real
- is the density that normalizes the distribution (default: 0.0334).
- MINImum-image
- BOXX=
 real
real BOXY=
BOXY= real
real
BOXZ= real
real END defines x,y,z
dimensions of a rectangular box for periodic boundary conditions
(default: no periodic boundary conditions).
END defines x,y,z
dimensions of a rectangular box for periodic boundary conditions
(default: no periodic boundary conditions).
- MODE=
 AVERage
AVERage CENTer
CENTer POINt
POINt
- provides alternate ways of computing angular distributions. AVERage computes distributions around each A-species atom within RADIus around ORIGin and averages them. CENTer computes the distribution around the geometric center of all A-species. POINt does it around the fixed ORIGin
- ORIGin=
 vector
vector
- is explained under MODE (default: (0 0 0)).
- OUTPut=
 filename
filename
- writes the radial distribution function to the specified file (default: OUTPUT).
- RADIus=
 real
real
- is explained under MODE (default: 0).
- RGRID=
 real
real
- is the interval between values of
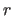 for which
for which 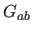 is
evaluated (default: 0.5).
is
evaluated (default: 0.5).
- RMAX=
 real
real
- provides a radial distribution
window upper limit for
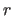 (default: 10.0).
(default: 10.0).
- RMIN=
 real
real
- provides a radial distribution window
lower limit for
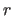 (default: 2.0).
(default: 2.0).
