| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Requirements Up: NCS Restraints Previous: NCS Restraints
Syntax
- NCS RESTraints {
 NCS-restraints-statement
NCS-restraints-statement } END
} END - is invoked from the main level of X-PLOR.
 NCS-restraints-statement
NCS-restraints-statement :==
:==-
- GROUp
- {
 ncs-restraints-group-statement
ncs-restraints-group-statement } END
adds a new group to the restraint NCS database.
} END
adds a new group to the restraint NCS database.
- INITialize
- erases the current restraint NCS database. If no restraints are specified, the energy flag NCS should be turned off as well.
 NCS-restraints-group-statement
NCS-restraints-group-statement :==
:==-
- EQUIvalence=
 selection
selection
- adds a new set of NCS-equivalent atoms within the group to the restraint NCS database.
- SIGB=
 real
real
- gives the target deviation
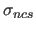 for B-factor restraints
(deviation from the average B of equivalent atoms) in Å
for B-factor restraints
(deviation from the average B of equivalent atoms) in Å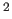 (default=2.0 Å
(default=2.0 Å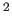 ).
).
- WEIGht=
 real
real
- gives the
effective energy constant for the positional
restraints in kcal mole
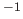 Å
Å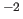 (default: 300.0
kcal mole
(default: 300.0
kcal mole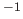 Å
Å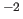 ).
).
- EQUIvalence=
Xplor-NIH 2025-11-07
