| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: CPU and Memory Requirements Up: Distance Geometry Previous: Test for the Correct
Regularization
X-PLOR can regularize coordinates that are
obtained by distance geometry embedding by minimizing
the coordinates against an effective energy term (DG) that represents
all upper and lower bounds in the distance geometry
bounds matrix. The bounds matrix has to be stored
in memory using the STOREBounds statement prior to
invoking the DG energy term. The atoms
that should be restrained have to be defined through the
SELEction statement. The DG term is defined as
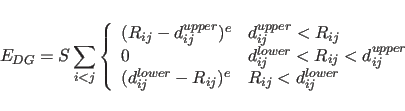 |
(37.1) |
The DG energy term is turned on by using the flag statement (see Section 4.5). Normally, all other energy terms should be turned off when using the DG term. However, for defining the bounds matrix initially, the other energy terms should be turned on, because bounds are computed only for the activated energy terms. Then, after storing the bounds matrix, all other energy terms should be turned off and the DG energy term turned on. Energy minimization, molecular dynamics, or any other energy calculation can be performed with the DG energy term.
Xplor-NIH 2025-11-07
