| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Requirements Up: Cartesian Coordinate Space Previous: Dynamics Restarts
Syntax of the Dynamics Verlet
Statement
The dynamics Verlet statement sets up various parameters for molecular dynamics
as well as Langevin dynamics and then executes the specified
number of steps.
- DYNAmics VERLet
- {
 dynamics-Verlet-statement
dynamics-Verlet-statement } END
is invoked from the main level of X-PLOR.
} END
is invoked from the main level of X-PLOR.
 dynamics-Verlet-statement
dynamics-Verlet-statement :==
:==-
- ASCIi=
 logical
logical
- writes a formatted (ASCII) coordinate
trajectory
file if ASCIi is TRUE;
if ASCIi is FALSE, writes an unformatted (binary) file.
In the former case,
a scale factor
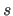 and an offset
and an offset 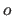 are applied to the
coordinates
are applied to the
coordinates 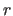 (i.e.,
(i.e.,
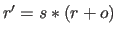 ); the result is converted to an
integer number and written to the
ASCII file using the specified format.
If a hexadecimal format is specified,
care should be taken to ensure that the
application of the scale factor
); the result is converted to an
integer number and written to the
ASCII file using the specified format.
If a hexadecimal format is specified,
care should be taken to ensure that the
application of the scale factor 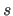 and the offset
and the offset 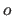 results in positive
coordinates
during
the whole course of the molecular dynamics simulation.
Otherwise, FORTRAN format runtime errors will occur.
results in positive
coordinates
during
the whole course of the molecular dynamics simulation.
Otherwise, FORTRAN format runtime errors will occur.
- CSEL=
 selection
selection
- selects atoms that are written to the coordinate trajectory file (default: all atoms except those fixed by the constraints fix statement).
- FINAltemp=
 real
real
- determines the target temperature for the velocity rescaling option (see ISCVel) (default: 298K).
- FIRSttemp=
 real
real
- assigns the temperature that determines the initial velocity (default: 0K).
- FORMat=
 string
string
- provides FORTRAN format for the ASCIi= TRUE option (default: 12Z.6, hexadecimal).
- IASVelocity=
 MAXWell
MAXWell UNIForm
UNIForm CURRent
CURRent
- determines the
mode of initial velocity assignments (default: zero velocities). - IEQFrq=
 integer
integer
- determines the frequency with which the velocities are rescaled (default: 0). It works in conjunction with FINAltemperature.
- ILBFrq=
 integer
integer
- updates Langevin boundary between normal and Langevin particles (default: 0). It works in conjunction with RBUF and ORIGin.
- IPRFrq=
 integer
integer
- prints energy statistics every IPRFrq steps if not equal to zero (default: 50).
- ISCVel=
 integer
integer
- determines the velocity rescale mode (default: 0).
- ISVFrq=
 integer
integer
- determines how often a restart file is written to disk (default: 0).
- NPRInt=
 integer
integer
- determines the frequency with which the energy is printed to standard output (default: 1, i.e., at every dynamics step).
- NSAVC=
 integer
integer
- determines the frequency with which the coordinates are written to the coordinate trajectory file (default: 0).
- NSAVV=
 integer
integer
- determines the frequency with which the velocities are written to the velocity trajectory file (default: 0).
- NSTEp=
 integer
integer
- determines the number of steps. In the case of a restart, the steps executed before writing the restart file are included in NSTEP (e.g., if NSTEP=3000, but 1000 steps were run before writing the restart file, the program will execute only 2000 steps) (default: 100).
- NTRFrq=
 integer
integer
- determines the frequency with which the center-of-mass motion is removed (default: 0).
- OFFSet=
 real
real
- provides offset
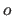 for the ASCIi=TRUE
option (default: 800).
for the ASCIi=TRUE
option (default: 800).
- ORIGin=
 vector
vector
- is the origin of the sphere of the Langevin boundary (default: 0,0,0).
- RBUF=
 real
real
- specifies the radius of the spherical boundary in a dynamics Verlet statement. Inside the boundary, the particles are “normal" molecular dynamics particles; outside they are propagated as Langevin particles (default: 0).
- RESTart=filename
- reads restart information from the named file and restarts the dynamics (default: no restart).
- SAVE=
 filename
filename
- specifies the name of the file to which the program periodically writes the restart information (default: none).
- SCALe=
 real
real
- provides a scale factor
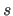 for the
ASCIi=TRUE option (default: 10000).
for the
ASCIi=TRUE option (default: 10000).
- TBATh=
 real
real
- specifies the temperature of the heatbath or the target temperature of Berendsen's temperature-coupling method.
- TCOUpling=
 logical
logical
- is a logical flag that turns on Berendsen's temperature-coupling method. This flag works in conjunction with TBATh (default: FALSE).
- TIMEstep=
 real
real
- assigns the time step
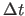 value for the finite-difference integration in psec
(default: 0.001 psec).
value for the finite-difference integration in psec
(default: 0.001 psec).
- TRAJectory=
 file
file
- provides the filename of the coordinate trajectory file (default: none).
- VASCii=
 logical
logical
- writes
a formatted (ASCII) velocity
trajectory file if VASCii is TRUE; if VASCii
is FALSE, an unformatted (binary) velocity
trajectory file is written.
In the former case,
a scale factor
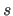 and an offset
and an offset 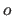 are applied to the
velocities
are applied to the
velocities 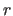 (i.e.,
(i.e.,
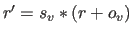 ); the result is converted to an
integer number and written to the
ASCII file using the specified format.
If a hexadecimal format is specified,
care should be taken to ensure that the
application of the scale factor
); the result is converted to an
integer number and written to the
ASCII file using the specified format.
If a hexadecimal format is specified,
care should be taken to ensure that the
application of the scale factor 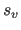 and the offset
and the offset 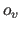 results in positive
velocities
during
the whole course of the molecular dynamics simulation.
Otherwise, FORTRAN format runtime errors will occur.
results in positive
velocities
during
the whole course of the molecular dynamics simulation.
Otherwise, FORTRAN format runtime errors will occur.
- VELOcity=
 filename
filename
- names the velocity trajectory file (default: none).
- VFORmat=
 string
string
- is the FORTRAN format of the velocity trajectory file for the VASCii= TRUE option (default: 12Z.6, hexadecimal).
- VOFFset=
 real
real
- provides an offset
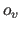 for the VASCii= TRUE
option (default: 300).
for the VASCii= TRUE
option (default: 300).
- VSCAle=
 real
real
- provides a scale factor
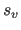 for the
VASCii=TRUE option (default: 10000).
for the
VASCii=TRUE option (default: 10000).
- VSEL=
 selection
selection
- selects atoms that are written to the velocity trajectory file (default: all atoms except those fixed by the constraints fix statement).
- ASCIi=
Xplor-NIH 2025-11-07
