| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Syntax Up: Molecular Replacement Previous: Requirements
Translation Search
X-PLOR performs a translation search by computing the target
function
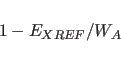 |
(19.6) |
The asymmetric unit of a translation function
is in general larger than the asymmetric unit of the unit cell.
For example,
in space group ![]() , the asymmetric unit for the translation function
is
, the asymmetric unit for the translation function
is ![]() ,
, ![]() ,
, ![]() , whereas the crystallographic
asymmetric unit is
, whereas the crystallographic
asymmetric unit is ![]() ,
, ![]() ,
,
![]() .
Hirshfeld (1968)
has defined the “Cheshire groups" for the 230 crystallographic
space groups. The unit cell of a particular Cheshire group corresponds
to an asymmetric unit of X-PLOR's translation function
(see Table 2 of the Hirshfeld paper). It is sometimes quite useful
to run a translation search on a very coarse lattice
that covers the complete unit cell (i.e.,
XGRId= 0 0.1 1, YGRId= 0. 0.1 1, ZGRId=0 0.1 1). Based on the
observed redundancies, one can then deduce other choices
for the asymmetric unit of the translation function not listed
in the Hirshfeld paper.
.
Hirshfeld (1968)
has defined the “Cheshire groups" for the 230 crystallographic
space groups. The unit cell of a particular Cheshire group corresponds
to an asymmetric unit of X-PLOR's translation function
(see Table 2 of the Hirshfeld paper). It is sometimes quite useful
to run a translation search on a very coarse lattice
that covers the complete unit cell (i.e.,
XGRId= 0 0.1 1, YGRId= 0. 0.1 1, ZGRId=0 0.1 1). Based on the
observed redundancies, one can then deduce other choices
for the asymmetric unit of the translation function not listed
in the Hirshfeld paper.
The output of the routine provides a complete listing of the computed
translation function, which is written to the specified
file. The format of this file is suitable for Mathematica.
In addition, a sorted list of a specified number
of highest peaks is
written to the standard output. A packing analysis is carried
out for each listed peak. The packing value consists
of the ratio of the molecular volume to the unit-cell volume;
it is expressed as a percentage.
The value of the translation function for the highest
peak is stored in the symbol $RESULT; the corresponding
packing function value is stored in the symbol $PACKING. The main
coordinate set is translated according to the highest translation
function value.
Subsections Xplor-NIH 2025-11-07
