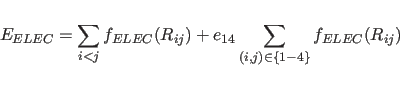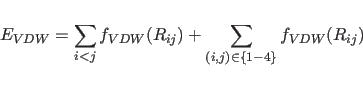| bit | Xplor-NIH | VMD-XPLOR |
|---|
|
| Xplor-NIH home Documentation |
Next: Crystallographic Symmetry Interactions Up: Nonbonded Energy Terms Previous: Electrostatic Function
Intramolecular Interactions
The intramolecular interaction energy is the summation of the individual
nonbonded interaction energies for pairs of atoms within the current
molecular
structure, e.g., a single molecule or a crystallographic asymmetric unit.
The summation extends over all pairs of atoms (
The computational time is further reduced
by introducing an approximation, storing the atomic pair
indices (![]() ) that satisfy
) that satisfy
![]() in a
list that is updated
only when any atom has moved more than the amount specified by
TOLErance. For both switched and shifted nonbonded
options (see Section 3.2.1),
the distance
in a
list that is updated
only when any atom has moved more than the amount specified by
TOLErance. For both switched and shifted nonbonded
options (see Section 3.2.1),
the distance ![]() at which the energy becomes
zero is given by
at which the energy becomes
zero is given by ![]() . Thus, the nonbonded energy
calculations become independent of the update frequencies if
. Thus, the nonbonded energy
calculations become independent of the update frequencies if
![]() .
.
There are a number of cases for nonbonded interactions that
must not be computed, e.g., interactions between covalently bonded
atoms. Covalently bonded exclusions are automatically generated
by X-PLOR using information about atom connectivity.
In addition, certain exclusions can be added manually by
the EXCLude statement, which is an atom statement
(see Section 3.1.1).
The NBXMod statement
(see Section 3.2.1)
has several options for automatically excluding
1-2, 1-2 and 1-3, and 1-2, 1-3,
and 1-4 interactions in the molecule. In the case of
NBXMod=![]() 5, the 1-4 interactions are treated in a special way.
The electrostatic 1-4 interactions are scaled by
5, the 1-4 interactions are treated in a special way.
The electrostatic 1-4 interactions are scaled by ![]() , and the
van der Waals interactions use a special 1-4 set of parameters for
, and the
van der Waals interactions use a special 1-4 set of parameters for
![]() and
and ![]() . In the case of
NBXMod
. In the case of
NBXMod![]() 5, 1-4 interactions are treated as normal nonbonded
interactions, and the second terms of the right-hand side of
Eqs. 4.17 and 4.18 become zero.
5, 1-4 interactions are treated as normal nonbonded
interactions, and the second terms of the right-hand side of
Eqs. 4.17 and 4.18 become zero.
Xplor-NIH 2025-11-07